
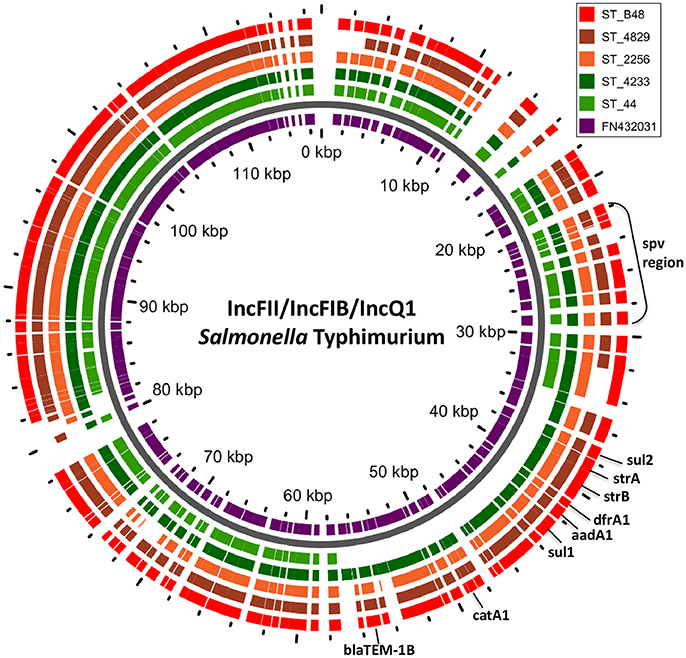
Primer pairs are evaluated against the PR 2 sequence database and you can test your own primers. We have developed in 2020 an interactive pr2-primers database focusing on the rRNA operon. Representation of the different taxonomic groups within PR 2. As an example, part of the ciliate annotation originate from the first EukRef workshop. EukRef has built bioinformatics pipelines that have been used during three workshops dedicated to specific taxonomic groups. One very important project in this respect is EukRef which has recently decided to merge its effort with PR 2. A number of metadata fields are available for many sequences, including geo-localisation, whether it originates from a culture or a natural sample, host type etc… The annotation of PR2 is performed by experts from each taxonomic groups. At present it contains about 184,000 sequences. Its aim is to provide a reference database of carefully annotated 18S rRNA sequences using eight unique taxonomic fields (from kingdom to species). The PR 2 sequence database was initiated in 2010 in the frame of the BioMarks project from work that had developed in the previous ten years in the Plankton Group of the Station Biologique of Roscoff. The origin of SARS‐CoV‐2 is a riddle: Meet the Twitter detectives who aim to solve it.The eukaryotic tree. Discovery of a novel coronavirus associated with the recent pneumonia outbreak in humans and its potential bat origin. A novel bat coronavirus closely related to SARS‐CoV‐2 contains natural insertions at the S1/S2 cleavage site of the spike protein. Viral metagenomics revealed Sendai virus and coronavirus infection of Malayan Pangolins (Manis javanica). A pneumonia outbreak associated with a new coronavirus of probable bat origin. (E) Phylogenetic tree of SARS‐GZ02, Pangolin/GX/2017, ZC45, ZXC21, RmYN02, RaTG13, and Pangolin/GD/2019 produced by CLUSTAL W based on the alignment of their genomes as in (B) RmYN02's nucleotides coding the PAA amino acids (surrounded by a red box) are aligned as mutations relative to ZC45 rather than insertions. (D) Pairwise comparisons of ZC45 (anchor) to ZXC21, RmYN02, RaTG13, and Pangolin/GD/2019. No PAA insertion is observed in RmYN02 in these comparisons. (C) Pairwise comparisons of RmYN02 (anchor) to RaTG13, ZC45, and ZXC21. The deletion characterizing RmYN02 at the S1/S2 junction appears to cause a split of the first nucleotide from the rest of the sequence coding for the PAA amino acids (CCT GCA GCG, surrounded by a red box). for comparison, with the exception of SARS‐CoV‐2. (B) Clustal W multiple sequence alignment of RmYN02 with the strains used in Zhou et al. No insertion in RmYN02 is visible on the contrary, a deletion splitting the nucleotides coding for PAA is observed. RmYN02's nucleotides coding the PAA amino acids (CCT GCA GCG) are surrounded by a red box. (A) Clustal W multiple sequence alignment of RmYN02 with the strains used in Zhou et al. BioEssays published by Wiley Periodicals LLC. Pangolin CoV MP789 RaTG13 RmYN02 SARS-CoV-2 furin cleavage site origin peer review. Because of their importance to inferring SARS-CoV-2's origin, we call for a careful reevaluation of RaTG13, MP789 and RmYN02 sequencing records and assembly methods. Also, our analysis of RaTG13 and RmYN02's metagenomic datasets found unexpected reads which could indicate possible contamination. We show that RmYN02/RacCSxxx instead of the claimed insertion carry a 6-nucleotide deletion in the region and that the 12-nucleotide insertion in SARS-CoV-2 remains unique among Sarbecoviruses. We specifically address the presence in RmYN02 and closely related RacCSxxx strains of a claimed natural PAA/PVA amino acid insertion at the S1/S2 junction of their spike protein at the same position where the PRRA insertion in SARS-CoV-2 has created a polybasic furin cleavage site. Their genomes have been used to support a natural origin of SARS-CoV-2 but after a close examination all of them exhibit several issues. RaTG13, MP789, and RmYN02 are the strains closest to SARS-CoV-2, and their existence came to light only after the start of the pandemic.


 0 kommentar(er)
0 kommentar(er)
